How do pathogenic bacteria survive during infection?
Bacterial responses to harmful compounds have been part of my research portfolio since my PhD where I investigated enzymes that protect bacterial cells from harmful interactions with sulfite. Studies of sulfite-oxidizing enzymes subsequently became a main focus of my research.
Building on this expertise in bacterial stress responses and metalloenzymes, we are now investigating enzymes that mediate resistance of H. influenzae to oxidative stress
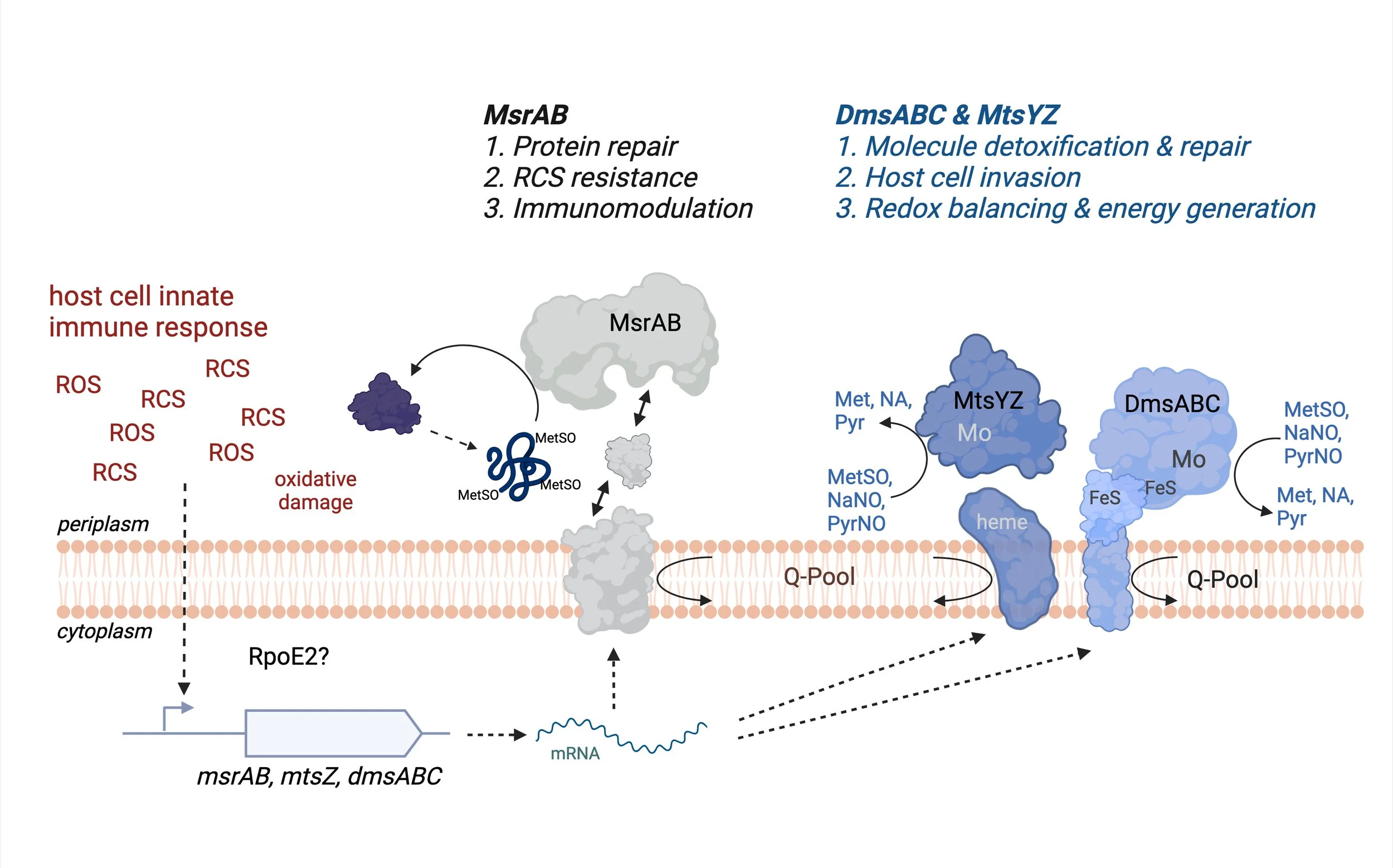
A particular focus have is resistance to reactive chlorine species (RCS) such as hypochlorite or N chlorotaurine (NCT), both of which easily damage sulfur compounds such as amino acids present in proteins by inducing formation of sulfoxides.
We have uncovered an RCS specific, extracellular stress response that uses two molybdenum enzymes, MtsZ and DmsABC (Frontiers in Microbiology 2016, 2021a, 2024) and a peptide methionine-sulfoxide reductase, MsrAB, to enhance bacterial fitness (ACS Infectious Disease, 2020, Antioxidants, 2022). While MsrAB is required for resistance to RCS killing and repairs damage to proteins located on the outside of the bacterial cell, i.e. in the bacterial cell envelope, MtsZ and DmsABC repair damage to nutrient molecules such as amino acids or vitamin precursors. Both MtsZ and DmsABC are essential for infection. Our extended characterization and crystal structure of the MtsZ enzyme also revealed how substrate specificity is achieved in this type of enzyme (JBC, 2021).
Expression of all three enzymes We also discovered that expression of these enzymes is induced not only by exposure to ROS such as hydrogen peroxide but also by hypochlorous acid, a reactive chlorine species (RCS) and is controlled by the first RCS responsive regulator in H. influenzae, the RpoE2 ECF sigma factor (Frontiers in Microbiology, 2021b, 2024).
Interested? We have student projects available for all our research areas. Please contact us for more information.
Images created with BioRender